|
|
3 years ago | |
|---|---|---|
| visualization | 3 years ago | |
| .gitignore | 3 years ago | |
| README.md | 3 years ago | |
| filtro.py | 3 years ago | |
| kenobi.cpp | 3 years ago | |
README.md
Closeness and Harmonic centrality over the IMDb Graph
Introduction
IMPORTANT: since Github does not render the math text, to properly read this README you have to clone the repo locally or install this extension that will render the math text.
This project is an exercise realized to implement a Social Network Analysis using the data of the Internet Movie Database (IMDb).
On this data we define an undirected graph G=(V,E) where
- the vertex V are the actor and the actress
- the non oriented edges in E links the actors and the actresses if they played together in a movie.
Interactive version here (it may take a few seconds to load)
The aim of the project was to build a social network over this graph and studying its centralities.
The first challenge was to filter the raw data downloaded from IMDb. One of the first (and funnier) problems was to delete all the actors that works in the Adult industry. They make a lot of movies together and this would have altered the results.
Then, the real challenge has come. We are working with a ton of actors, a brute force approach would have required years to compile: an efficient algorithm was necessary
Understanding the data
We are taking the data from the official IMDB dataset: https://datasets.imdbws.com/
In particolar we're interest in 3 files
title.basics.tsvtitle.principals.tsvname.basics.tsv
Let's have a closer look to this 3 files:
title.basics.tsv.gz
Contains the following information for titles:
- tconst (string) - alphanumeric unique identifier of the title
- titleType (string) – the type/format of the title (e.g. movie, short, tvseries, tvepisode, video, etc)
- primaryTitle (string) – the more popular title / the title used by the filmmakers on promotional materials at the point of release
- originalTitle (string) - original title, in the original language
- isAdult (boolean) - 0: non-adult title; 1: adult title
- startYear (YYYY) – represents the release year of a title. In the case of TV Series, it is the series start year
- endYear (YYYY) – TV Series end year. ‘\N’ for all other title types
- runtimeMinutes – primary runtime of the title, in minutes
- genres (string array) – includes up to three genres associated with the title
title.principals.tsv.gz
Contains the principal cast/crew for titles
- tconst (string) - alphanumeric unique identifier of the title
- ordering (integer) – a number to uniquely identify rows for a given titleId
- nconst (string) - alphanumeric unique identifier of the name/person
- category (string) - the category of job that person was in
- job (string) - the specific job title if applicable, else '\N'
- characters (string) - the name of the character played if applicable, else '\N'
name.basics.tsv.gz
Contains the following information for names:
- nconst (string) - alphanumeric unique identifier of the name/person
- primaryName (string)– name by which the person is most often credited
- birthYear – in YYYY format
- deathYear – in YYYY format if applicable, else '\N'
- primaryProfession (array of strings)– the top-3 professions of the person
- knownForTitles (array of tconsts) – titles the person is known for
Filtering
All This section refers to what's inside the file filtro.py
Now that we have downloaded all the files from the dataset, we have to filter them and modify them in order to easily work with them.
name.basics.tsv
For this file we only need the following columns
nconstprimaryTitleprimaryProfession
Since all the actors starts with the string nm0 we can remove it to clean the output. Furthermore a lot of actors/actresses do more than one job (director etc..). To avoid excluding important actors we consider all the ones that have the string actor/actress in their profession. In this way, both someone who is classified as actor or as actor, director is taken into consideration
Then we can generate the final filtered file Attori.txt that has only two columns: nconst and primaryName
title.basics.tsv.gz
For this file we only need the following columns
tconstprimaryTitleisAdulttitleType
Since all the movies starts with the string t0 we can remove it to clean the output. In this case, we also want to remove all the movies for adults.
There is a lot of junk in IMDb. To avoid dealing with un-useful data, we are considering all the non-adult movies in this whitelist
movietvSeriestvMovietvMiniSeries
Why this in particolar? Benefits on the computational cost. There are (really) a lot of single episodes listed in IMDb: to remove them without loosing the most important relations, we only consider the category tvSeries. This category list a TV-Series as a single element, not divided in multiple episodes. In this way we will loose some of the relations with minor actors that may appear in just a few episodes. But we will have preserved the relations between the protagonists of the show.
Then we can generate the final filtered file FilmFiltrati.txt that has only two columns: tconst and primaryTitle
title.principals.tsv
For this file we only need the following columns
tconstnconstcategory
As before, we clean the output removing unnecessary strings. Then we create an array of unique actor ids (nconst) and an array of how may times they appear (counts). This will give us the number of movies they appear in. And here it comes the core of this filtering. We define at the start of the algorithm a constant MIN_MOVIES. This integer is the minimum number of movies that an actor has to have done in his carrier to be considered in this graph. The reason to do that it's purely computational. If I have to consider all actors the time for the code to compile is in the year(s)'s order, that's not good. We are making an approximation: if an actor has less then a reasonable (42, as an example) number of movies made in his carrier, there is an high probability that he/she has an important role in our graph during the computation of the centralities.
Notice that we are only selecting actors and actresses that have at least a relation.
At the end, we can finally generate the file Relazioni.txt containing the columns tconst and nconst
Understanding the code
Now that we have understood the python code, let's start with the core of the algorithm, written in C++
Data structures to work with
In this case we are working with two simple struct for the classes Film and Actor
struct Film {
string name;
vector<int> actor_indicies;
};
struct Actor {
string name;
vector<int> film_indices;
};
Then we need two dictionaries build like this
map<int, Actor> A; // Dictionary {actor_id (key): Actor (value)}
map<int, Film> F; // Dictionary {film_id (key): Film (value)}
The comments explain everything needed
Data Read
This section refers to the function DataRead()
void DataRead()
{
ifstream actors("data/Attori.txt");
ifstream movies("data/FilmFiltrati.txt");
string s,t;
const string space /* the final frontier */ = "\t";
for (int i = 1; getline(actors,s); i++)
{
if (s.empty())
continue;
try {
Actor TmpObj;
int id = stoi(s.substr(0, s.find(space)));
TmpObj.name = s.substr(s.find(space)+1);
A[id] = TmpObj; // Matlab/Python notation, works with C++17
if (id > MAX_ACTOR_ID)
MAX_ACTOR_ID = id;
} catch (...) {
cout << "Could not read the line " << i << " of Actors file" << endl;
}
}
for (int i = 1; getline(movies,t); i++)
{
if (t.empty())
continue;
try{
Film TmpObj;
int id = stoi(t.substr(0, t.find(space)));
TmpObj.name = t.substr(t.find(space)+1);
F[id] = TmpObj;
} catch (...) {
cout << "Could not read the line " << i << " of Film file" << endl;
}
}
}
We are considering the files Attori.txt and FilmFiltrati.txt, we don't need the relations one for now. Once that we have read this two files, we loop on each one brutally filling the two dictionaries created before. If a line is empty, we skip it.
Building the Graph
This section refers to the function BuildGraph()
void BuildGraph()
{
ifstream relations("data/Relazioni.txt");
string s;
const string space = "\t";
for (int i=1; getline(relations,s); i++){
if (s.empty())
continue;
try {
int id_film = stoi(s.substr(0, s.find(space)));
int id_attore = stoi(s.substr(s.find(space)+1));
if (A.count(id_attore) && F.count(id_film)) { // Exclude movies and actors filtered
A[id_attore].film_indices.push_back(id_film);
F[id_film].actor_indicies.push_back(id_attore);
}
} catch (...) {
cout << "Could not read the line " << i << " of Releations file" << endl;
}
}
}
In this function, we only use the file Relazioni.txt. As done before, we loop on all the elements of this file, creating
id_film: index key of each movieid_attore: index key of each actor
If both exists then we update le list of indices of movies that the actor of that id played in. In the same way, we updated the list of indices of actors that played in the movies with that id.
Closeness Centrality
That's where I tried to experiment a little bit. The original idea to optimize the algorithm was to take a uniformly random subset of actors. This method has a problem: no matter how smart you take this random subset, you are going to exclude some important actors. And I would never want to exclude Ewan McGregor from something!
So I found this paper and I decided that this was the way to go
The problem
Given a connected graph G = (V, E), the closeness centrality of a vertex v is defined as
C(v) = \frac{n-1}{\displaystyle \sum_{\omega \in V} d(v,w)} The idea behind this definition is that a central node should be very efficient in spreading information to all other nodes: for this reason, a node is central if the average number of links needed to reach another node is small.
This measure is widely used in the analysis of real-world complex networks, and the problem of selecting the k most central vertices has been deeply analysed in the last decade. However, this problem is computationally not easy, especially for large networks.
This paper proposes a new algorithm that here is implemented to compute the most central actors in the IMDB collaboration network, where two actors are linked if they played together in a movie.
In order to compute the k vertices with largest closeness, the textbook algorithm computes
c(v) for each v and returns the k largest found values. The main bottleneck of this approach
is the computation of d(v, w) for each pair of vertices v and w (that is, solving the All
Pairs Shortest Paths or APSP problem). This can be done in two ways: either by using fast
matrix multiplication, in time O(n^{2.373} \log n) [Zwick 2002; Williams 2012], or by performing a breadth-first search (in short, BFS) from each vertex v \in V , in time O(mn), where n = |V| and m = |E|. Usually, the BFS approach is preferred because the other approach contains big constants hidden in the O notation, and because real-world networks are usually sparse, that is, m is not much bigger than n. However, also this approach is too time-consuming if the input graph is very big
Preliminaries
In a connected graph, the farness of a node v in a graph G = (V,E) is
f(v) = \frac{1}{n-1} \displaystyle \sum_{\omega \in V} d(v,w)and the closeness centrality of v is 1/f(v) . In the disconnected case, the most natural generalization would be
f(v) = \frac{1}{r(v)-1}\displaystyle \sum_{\omega \in R(v)} d(v,w) and c(v)=1/f(v), where R(v) is the set of vertices reachable from v, and r(v) = |R(v)|.
But there is a problem: if v has only one neighbor w at distance 1, and w has out-degree 0, then v becomes very central according to this measure, even if v is intuitively peripheral. For this reason, we consider the following generalization, which is quite established in the literature [Lin 1976; Wasserman and Faust 1994; Boldi and Vigna 2013; 2014; Olsen et al. 2014]:
f(v) = \frac{n-1}{(r(v)-1)^2}\displaystyle \sum_{\omega \in R(v)} d(v,w) \qquad \qquad c(v)= \frac{1}{f(v)} If a vertex v has (out)degree 0, the previous fraction becomes \frac{0}{0} : in this case, the closeness of v is set to 0
The algorithm
In this section, we describe our new approach for computing the k nodes with maximum closeness (equivalently, the k nodes with minimum farness, where the farness f(v) of a vertex is 1/c(v) as defined before.
The basic idea is to keep track of a lower bound on the farness of each node, and to skip the analysis of a vertex v if this lower bound implies that v is not in the top k.
-
Firstly we compute the farness of the first
kvertices and save them in vectortop_actors -
Then, for all the next vertices, we define a lower bound
$$ \frac{n-1}{(n-1)^2} (\sigma_{d-1} + n_d \cdot dwhere
\sigmais the partial sum. This lower bound is updated each time that we move to another level of exploration during the BFS. In this way, if at a change of level in the BFS the lower bound of the vertex that we are computing is bigger than the k-th element oftop_actors, we can skip it. Remember that the bigger the farness the lower the closeness. The idea is that: if at this level, it's already that bad, it can't improve during the remaining part of the BFS. So there is no reason to continue the computingInstead, if at every level this lower bound is smaller than the k-th element of
top_actors(which is the element with the biggest farness computed till now) it means that we have to add it to the vector and remove the last one.I choose this particular lower bound because it's the worst case possible. When we are at a new level
dof the exploration we have already computed the sum of the formula up to the leveld-1. And now the worst case at this level is that our vertex is connected to all the other vertices at leveld(that aren_d).
An interactive graph showing the relation between the actors with highest closeness centrality
Harmonic Centrality
The algorithm described before can be easy applied to the harmonic centrality, defined as
h(v) = \sum_{w \in V} \frac{1}{d(v,w)} The main difference here is that we don't have a farness (where small farness implied bigger centrality). Then we won't need a lower bound either. Since the biggest the number is the higher is the centrality we have to adapt the algorithm.
Instead of a lower bound, we need an upper bound such that
h(v) \leq U_B (v) \leq h(w) We can easily define considering the worst case that could happen at each state:
U_b (v) = \sigma_{d-1} + \frac{n_d}{d} + \frac{n - r - n_d}{d+1}Why this? We are at the level d of our exploration, so we already know the partial sum \sigma_{d-1}. The worst case here in this level were we are connected to all the other nodes so we add the other two factors \frac{n_d}{d} + \frac{n - r - n_d}{d+1}
Then the algorithm works with the same top-k philosophy, just with an upper bound instead of a lower bound
An interactive graph showing the relation between the actors with highest closeness centrality
Benchmarks
Tested on Razer Blade 15 (2018) with an i7-8750H (6 core, 12 thread) and 16GB of DDR4 2666MHz RAM. The algorithm is taking full advantage of all 12 threads
| MIN_ACTORS | k | Time for filtering | Time to compile |
|---|---|---|---|
| 42 | 100 | 1m 30s | 3m 48s |
| 31 | 100 | 1m 44s | 8m 14s |
| 20 | 100 | 2m 4s | 19m 34s |
| 15 | 100 | 2m 1s | 37m 34s |
| 5 | 100 | 2m 10s | 2h 52m 57s |
How the files changes in relation to MIN_ACTORS
| MIN_ACTORS | Attori.txt elements | FilmFiltrati.txt elements | Relazioni.txt elements |
|---|---|---|---|
| 42 | 7921 | 266337 | 545848 |
| 31 | 13632 | 325087 | 748580 |
| 20 | 26337 | 394630 | 1056544 |
| 15 | 37955 | 431792 | 1251717 |
| 5 | 126771 | 547306 | 1949325 |
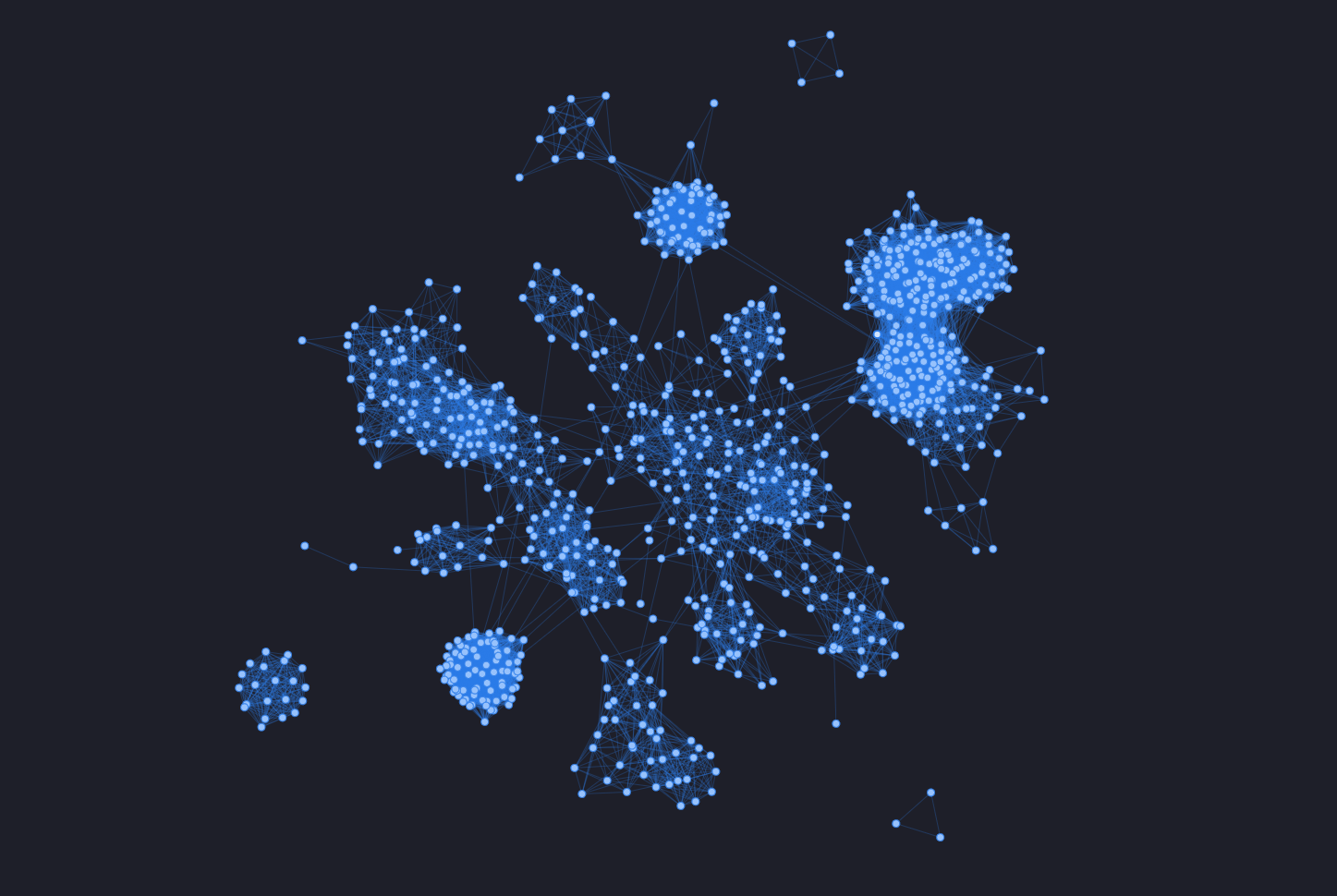
Visualization
One of the funniest part of working with graph is visualizing them, isn't it? Thanks to the python library pyvis I was able to generate an html file with an interactive version of the IMDb interactions graph.
To avoid creating a heavy and buggy webpage I have taken into consideration only a small set of actors. To do that I used the already working python script described before, considering only the actors with at least 100 movies made in their carrier.
This created a very interesting graph: there are some very strong neighborhood almost isolated from the rest. One explanation can be found in the Bollywood community. A lot of people making a lot of movies only for the indian market. This leads to the creation of a neighborhood strongly connected (not in the math way) but isolated form the other community, as the hollywood one as an example.